New to DrugBank?
Check out our Clinical Trial and Rare Disease data package in our Data Library .
Clinical trials represent a valuable source of information for drug discovery and repurposing efforts. Reviewing past trials for a molecule of interest, or molecules similar to one under study, can help researchers understand the potential outcomes of future trials. In the case of drug repurposing, it is essential to understand previous populations a drug has been tested on. This can not only confirm that the drug is truly being repurposed for a novel patient population but also provide useful information on previous trial outcomes to inform future trial design.
One of the most widely used sources for clinical trial data is clinicaltrials.gov maintained by the NIH. Investigators can submit valuable information, including metadata (e.g., dates, study design, etc.), to the database. Unfortunately, this self-reporting mechanism relies heavily on unstructured text fields and largely lacks quality control. As a result, users often have to spend significant time and resources to extract, clean, structure, and store the data to extract meaning and value from it.
DrugBank regularly imports this clinical trial data and, by cleaning, structuring, and linking it to other parts of our knowledge base, offers a highly usable dataset. This article explores the DrugBank clinical trial data and provides examples of useful queries to answer particular questions.
Check out our Clinical Trial and Rare Disease data package in our Data Library .
Faster drug discovery starts here.
The clinical trials dataset is centered on our clinical_trials table but also contains several additional supporting tables to provide more details for a given trial. Often, trial patients will be randomized into groups (“arm groups”) to receive specific treatments (“interventions”). Within DrugBank, the clinical_trial_arm_groups table captures these patient groupings for each trial, including what type the group refers to:
| Arm group "kind" | Description |
|---|---|
| experimental | The specific intervention to be evaluated in the study |
| active_comparator | An existing treatment known to be effective to which an experimental treatment is being compared |
| placebo_comparator | Placebo; an intervention that mimics the experimental intervention but has no therapeutic value |
| sham_comparator | A procedure or device that appears similar to the experimental procedure or device but yields no therapeutic value |
| no_intervention | No intervention is applied |
| other | Any intervention not classified above |
| unknown | Intervention data is missing or otherwise invalid |
The specific interventions are tied to the trial itself (through the ‘trial_id’ column of the clinical_trial_interventions table) and to individual arm groups (through the clinical_trial_intervention_arm_groups mapping table). Interventions are mapped to DrugBank drugs and existing products, where appropriate, through the clinical_trial_interventions_drugs and clinical_trial_interventions_products tables.
Other tables within the clinical trials dataset will be explored and explained through the sample queries outlined below. The majority of these tables are straightforward in both their meaning and interpretation.
As an example, let’s consider retrieving all phase I and II trials that started from April 2020 until the end of 2022, during the height of the COVID-19 pandemic:
SELECT ct.identifier, ct.title, ct.start_date, ctp.phase FROM clinical_trials ct JOIN clinical_trial_phases ctp ON ct.identifier=ctp.trial_id WHERE ct.start_date BETWEEN '2020-04-01' AND '2022-12-31' AND ctp.phase IN (1, 2) ORDER BY ct.start_date;Copy to clipboard
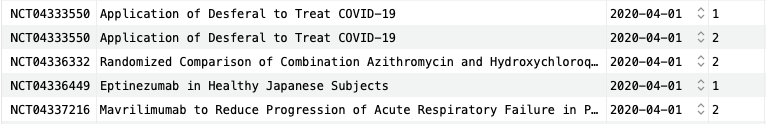
Five rows are shown here, including two trials in phase I and three trials in phase II. Already, we can see two trials with "COVID-19" in the title and, although it is cut off, the third and fifth trials are also related to COVID-19.
This query shows a general pattern for working with clinical trial data in DrugBank, wherein the main clinical trial data is joined to additional specific data as needed. Additional filtering for attributes such as the start or end date of a trial, the trial phase, or other attributes can help make queries more specific.
This query involves only a single join between the clinical_trials and clinical_trial_phases tables. The main join column within the clinical trials dataset is the trial ID itself, which is found in the ‘identifier’ column in the main table and as ‘trial_id’ in all other tables.
Although there is a lot of value in the individual clinical trial tables, more insights can be obtained by merging with the larger DrugBank knowledge base. One way to do this is to rely on condition mapping, which ties the condition(s) under study to our larger conditions dataset. As an example, let’s consider finding all trials for a given condition, in this case, diabetes mellitus (DBCOND0027886):
SELECT ct.identifier, ct.title, ctp.phase, cte.gender, cte.healthy_volunteers, cte.criteria FROM clinical_trials ct JOIN clinical_trial_phases ctp ON ct.identifier=ctp.trial_id JOIN clinical_trial_conditions ctc ON ct.identifier=ctc.trial_id JOIN conditions c ON ctc.condition_id=c.id JOIN clinical_trial_eligibilities cte ON ct.identifier=cte.trial_id WHERE c.drugbank_id = 'DBCOND0027886';Copy to clipboard
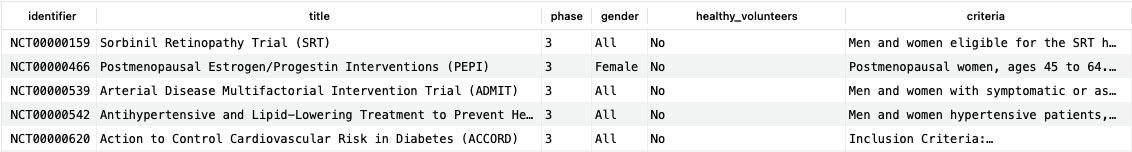
This query returns just over 500 rows at the time of writing, indicating that there have been a substantial number of trials for this condition; the first five are shown here.
This query demonstrates an important pattern: the filtering of trial results by a disease or condition under study. Although we limited this filtering here to a single condition, it is possible to extend this to a series of conditions, or to conditions based on their mapping to an external code such as ICD-10. In addition, we are joining the recently added trial eligibilities data. The clinical_trial_eligibilities table contains the inclusion and exclusion criteria for each trial; this can be used to evaluate how recruiting was carried out for previous trials. Such information on (un)successful trial designs can be very useful when designing future trials in order to maximize the chance of success.
Here, we are joining an additional supporting table, clinical_trial_eligibilities, to the main clinical_trials table, again using the trial identifier as above. To include conditions data, we first join the clinical_trial_conditions mapping table, which acts as a bridge between clinical trials and conditions. This table can be joined by using the trial identifier. Then, to join the conditions table, we will join this to the clinical_trial_conditions table by equating the ‘condition_id’ in this table to the ‘id’ in the conditions table.
Note: There is a similarly named clinical_trial_browse_conditions table, as well as a clinical_trial_browse_interventions table, which mirrors the clinical_trial_interventions table. These are based solely on the automated extraction of concepts by clinicaltrials.gov and are less reliable.
Similar to the previous example focussing on (a) specific condition(s), it is also possible to focus on trials which include a specific drug as an intervention:
SELECT ct.identifier, ctp.phase, c.drugbank_id, c.title, ct.start_date FROM clinical_trials ct JOIN clinical_trial_phases ctp ON ct.identifier=ctp.trial_id JOIN clinical_trial_conditions ctc ON ct.identifier=ctc.trial_id JOIN conditions c ON ctc.condition_id=c.id JOIN clinical_trial_interventions cti ON ct.identifier=cti.trial_id JOIN clinical_trial_interventions_drugs ctid ON cti.id=ctid.intervention_id JOIN drugs d ON ctid.drug_id=d.id WHERE d.drugbank_id = 'DB06273' ORDER BY ct.start_date DESC;Copy to clipboard
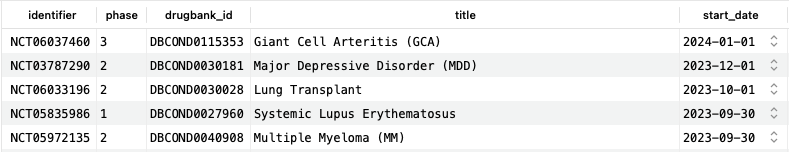
This query shows trials for which the IL-6 receptor inhibiting monoclonal antibody tocilizumab (DB06273) is used as an intervention, organized by date. At the time of writing, there are 670 such trials, five of which are shown here, and some have a start date past the date of writing, indicating future starts.
Much like the above example, this pattern is important for those looking to understand the trials for a specific drug or list of drugs. Although we chose only to show the associated condition under investigation, other information might be of interest. For example, by including the arm groups data, it would be possible to investigate further whether the drug was being studied directly or being used as a comparator.
The clinical_trial_phases and conditions tables are joined as shown previously. As there is no direct connection between the clinical_trials and drugs tables, we must use intermediate tables related to the trial interventions. To this end, we need 3 steps:
As drugs and conditions are two major connecting data types within DrugBank, it is relatively easy to use them to connect other datasets. These connections can allow for powerful queries capable of answering realistic questions. As an example, finding conditions under study in clinical trials for which there is no drug currently indicated:
SELECT c.id, c.drugbank_id, c.title, COUNT(*) AS count FROM clinical_trials ct JOIN clinical_trial_conditions ctc ON ct.identifier=ctc.trial_id JOIN conditions c ON ctc.condition_id=c.id WHERE c.id NOT IN ( SELECT DISTINCT c.id FROM structured_indications si LEFT JOIN indication_conditions icp ON ( si.id=icp.indication_id AND (icp.relationship = 'for_condition' OR icp.relationship = 'associated_condition') ) LEFT JOIN conditions c ON (icp.condition_id=c.id) WHERE c.title != '' ) GROUP BY c.title ORDER BY count DESC;Copy to clipboard
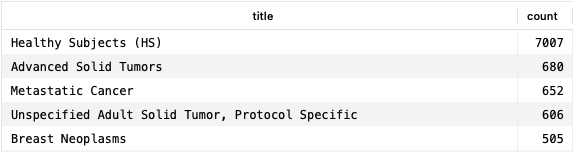
This query provides conditions fitting these criteria, and the number of times they appear in the clinical trials dataset. As a condition for which we would not expect an indication, “Healthy Subjects” makes a lot of sense.
This query is a good example of a more general pattern that can be applied across basically any dataset within DrugBank; namely, filtering the trials dataset by another criteria. Here, we have chosen indications, but a similar approach could be used to, for example, find trials relating to drugs with a specific target, adverse effect, etc. Leveraging these connections to apply specific filters is a powerful way to explore subsets of clinical trials.
The clinical_trial_phases and conditions tables are joined, as shown previously. This query is more complex than those shown previously as it uses a subquery. Within the subquery, we first join the structured_indications table to the indication_conditions table using the ‘id’ in the former and the ‘indication_id’ in the latter. There is an additional criterion used in this join. Within the indications dataset, conditions can be used in a variety of ways such as designating the condition to be treated or by capturing additional information about the intended treatment population. Here, we specify that the condition must be related to what is being treated (either “for_condition” or “associated_condition”). We then join the result to the conditions table by equating the ‘condition_id’ to the ‘id’ in the conditions table itself. By selecting just the resulting DBCOND identifiers, we can use this list to filter the trial conditions within the main query.
Clinical trials information can provide a wealth of information on previous investigations into drugs and disease areas. Although clinicaltrials.gov provides access to these data, the entries are often messy and difficult to work with. DrugBank offers a cleaned and structured version of these data, which are also connected to the entire rest of our knowledgebase. By leveraging these connections, it is possible to query subsets of trials relating to some defined criteria and quickly answer complex questions around drugs and conditions under study.