New to DrugBank?
Check out our Clinical Trial and Rare Disease data package in our Data Library .
A significant proportion of the global population (3.5-5.9%) live with rare conditions that lack effective treatment options. Interest in rare disease research has increased in recent years, driven partly by government programs incentivizing research in these areas. To assist rare disease researchers, we have added new data to our knowledgebase in this area.
The question of what precisely defines a rare disease is difficult to answer, as it largely depends on who you ask. According to Eurodis and Rare Disease UK, the European Union currently defines a rare disease as affecting fewer than 1 in 2,000 individuals. Conversely, the United States currently defines a rare disease as any disease that affects fewer than 200,000 people in the US. Based on the current US population, this works out to be roughly 1 in 1,700 individuals.
Hence, although the exact threshold chosen is important, the list of rare diseases obtained is likely very similar across jurisdictions and definitions. Currently, DrugBank follows the US definition of a rare disease, as our dataset is based on US orphan drug designations.
Check out our Clinical Trial and Rare Disease data package in our Data Library .
Faster drug discovery starts here.
The US grants orphan drug designations to drugs or biological products intended to prevent, diagnose, or treat a rare disease. As such, each designation includes one or more drugs but only a single rare disease. Within DrugBank, each designation is a structured entry that maps to one or more drugs and a single condition. As an example, let’s find all the current orphan designations for lenalidomide (DB00480):
SELECT d.drugbank_id, d.name, ofd.orphan_designation, c.drugbank_id, c.title, ofd.designation_date, ofd.designation_status FROM drugs d JOIN orphan_designation_drugs odd ON d.id=odd.drug_id JOIN orphan_designations od ON odd.orphan_designation_id=od.id JOIN orphan_fda_designations ofd ON od.id=ofd.orphan_designation_id JOIN conditions c ON od.condition_id=c.id WHERE d.drugbank_id = 'DB00480' ORDER BY ofd.designation_date;Copy to clipboard
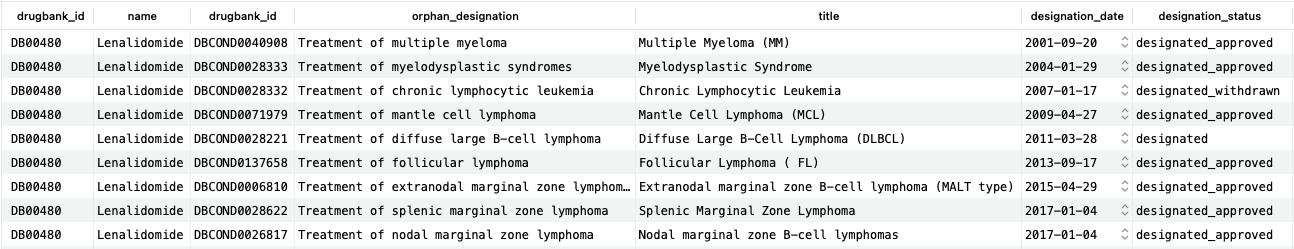
As we can see, lenalidomide has nine FDA orphan designations for various types of blood cancers. Although each entry has more associated information, we include only a subset here for clarity. The “designation_status” column shows the current status of the designation, including whether there was subsequently an approved product or if the designation was withdrawn.
One aspect that may be confusing is the existence of both an orphan_designations and an orphan_fda_designations table (e.g., in the below query). Each entry in the orphan_designations table is a generic entry capturing some basic information about the designation, while the orphan_fda_designations table lists more information specific to the FDA’s designations. Future improvements may include designations from other organizations, which would then follow a similar structure and could necessitate additional tables specific to the new organizations.
Although straightforward, this query represents a good starting point for other queries relating to the rare disease data. As an example, though we focussed on designations for a single drug, it is straightforward to modify this query to instead find all the designations associated with a specific condition.
Here, we begin by joining the drugs table to the orphan_designation_drugs table using the drug ‘id’ and the orphan drug ‘drug_id’ columns. This table serves to map between orphan designations and drug identifiers. Next, we join this to the orphan_designations table by relating the ‘orphan_designation_id’ to the ‘id’ in the main table. We then join the orphan_fda_designations table, once again using the ‘orphan_designation_id.’ This table contains most of the specific information for the designations in question. Finally, we join to the conditions table, this time by equating the condition ‘id’ to the ‘condition_id’ within the main orphan_designations table.
As is often the case, it is the connectivity between data points, more than the data points themselves, that provides the most value when working with DrugBank. As the orphan designations are linked to both drugs and conditions, it is possible to obtain much more context around them. For example, let’s consider finding existing indicated drugs for the conditions for which lenalidomide has received its orphan designations:
SELECT c_rd.drugbank_id, c_rd.title, GROUP_CONCAT(DISTINCT Drugs SEPARATOR '; ') AS 'Indicated Drugs', ofd.designation_date, ofd.designation_status FROM drugs d_rd JOIN orphan_designation_drugs odd ON d_rd.id=odd.drug_id JOIN orphan_designations od ON odd.orphan_designation_id=od.id JOIN orphan_fda_designations ofd ON od.id=ofd.orphan_designation_id JOIN conditions c_rd ON od.condition_id=c_rd.id LEFT JOIN ( SELECT ic_f.condition_id AS 'condition_id', d_pi.name AS 'Drugs' FROM structured_indications si_f JOIN indication_conditions ic_f ON ( si_f.id=ic_f.indication_id AND (ic_f.relationship = 'for_condition' OR ic_f.relationship = 'associated_condition') ) LEFT JOIN drugs d_pi ON si_f.drug_id=d_pi.id UNION SELECT ic_s.condition_id AS 'condition_id', d_se.name AS 'Drugs' FROM structured_indications si_s JOIN indication_conditions ic_s ON ( si_s.id=ic_s.indication_id AND (ic_s.relationship = 'for_condition' OR ic_s.relationship = 'associated_condition') ) LEFT JOIN indication_drugs id ON si_s.id=id.indication_id LEFT JOIN drugs d_se ON id.drug_id=d_se.id ORDER BY Drugs ) AS indicated_drugs USING(condition_id) WHERE d_rd.drugbank_id = 'DB00480' GROUP BY ofd.orphan_designation_id ORDER BY ofd.designation_date;Copy to clipboard
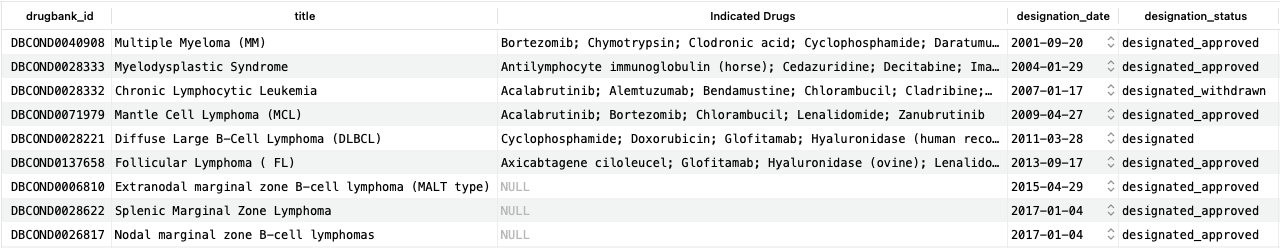
As we can see, for the first six designations, multiple drugs are already indicated to treat them, while for the final three, there are none.
When considering a research area to pursue, it is often useful to understand how crowded with other researchers, drugs, and treatments that particular area is. To gain a deeper understanding of the competitive landscape and to help determine which areas may benefit from so-called "first-in-class" versus "best-in-class" therapies, we can execute a query to investigate existing indications for a given condition. Similarly, one could extend beyond examining indications to incorporate existing market products to determine market saturation. The highly relational and connected nature of DrugBank data means it is easy to connect the orphan designations to any other dataset to gain additional insight.
Here, the first several joined tables are identical to those in the previous query. The main difference comes from the large subquery inserted afterwards using a LEFT JOIN. In our indications dataset, there is always at least one main drug in the structured_indications table and zero or more associated drugs in a separate indication_drugs table. To ensure we can capture only the unique entries across both tables, we implement a UNION of the two tables, capturing both the 'condition_id' and the actual drug names; the 'condition_id' is important for the actual joining operation to the main query body. In the first part of the subquery, we join the structured_indications table to the indication_conditions table. Here, we are joining on the indication ID but with the additional constraint that the condition in question needs to be either the primary (‘for_condition’) or secondary (‘associated_condition’) condition being treated or managed by the drug(s). The second part of the subquery is essentially the same, but we additionally join the indication_drugs table to capture those additional drugs in multi-drug indications. Finally, in both cases, we join back to the drugs table by equating the ‘drug_id’ to the ‘id’ of the drug. The result of the subquery is then joined to the main query by equating the ‘condition_id’ of the orphan designation to that of the existing indication.
The two main connections to clinical trials within DrugBank are drugs and conditions. These are also the main connections to orphan drug designations. As such, it is fairly straightforward to find clinical trials related to a designation. This could be done by matching the designation drug(s) to those used in trials, by matching the designation condition, or both. The closest match is achieved by filtering on both drug(s) and condition; an example of a query that does this for lenalidomide (DB00480) is shown below:
SELECT DISTINCT ofd.orphan_designation, ct.identifier, ct.title AS 'trial title', c.title AS 'condition name', ct.start_date FROM orphan_designations od JOIN orphan_designation_drugs odd ON od.id=odd.orphan_designation_id JOIN orphan_fda_designations ofd ON od.id=ofd.orphan_designation_id JOIN clinical_trial_interventions_drugs ctid ON odd.drug_id=ctid.drug_id JOIN clinical_trial_interventions cti ON ctid.intervention_id=cti.id JOIN clinical_trial_intervention_arm_groups ctiag ON cti.id=ctiag.intervention_id JOIN clinical_trial_arm_groups ctag ON ctiag.arm_group_id=ctag.id JOIN clinical_trials ct ON cti.trial_id=ct.identifier JOIN clinical_trial_conditions ctc ON ( ct.identifier=ctc.trial_id AND ctc.condition_id=od.condition_id ) JOIN conditions c ON od.condition_id=c.id WHERE odd.drug_id = 480 AND ctag.kind = 'experimental';Copy to clipboard
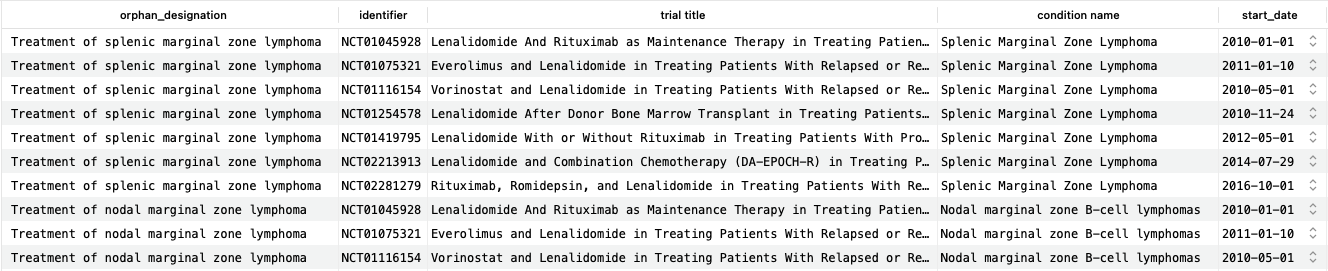
This query returns just over 500 matching rows (corresponding to 500 unique trial identifiers), the first 10 of which are shown here. As we can see, the returned trials include lenalidomide (often in combination with other drugs) as an intervention and match the condition for the designation. It is possible to further restrict the trials to those that use only lenalidomide, but for simplicity we will not explore this here.
Queries such as these are used to filter DrugBank's clinical trial data for the purpose of locating closely matching orphan drug designations. These trials can then be investigated to understand aspects such as trial design, eligibility criteria, and outcome. This can help in designing future trials or just provide useful context to understand the existing research landscape in a particular disease.
Here, we are joining the various tables related to orphan designations as described previously. Many of the join operations to clinical trial tables were also described in depth in another article. What is new here are the clinical_trial_intervention_arm_groups and clinical_trial_arm_groups tables. The first is joined by equating the ‘intervention_id’ in the table to the ‘id’ of the clinial_trial_interventions table. Then, the clinical_trial_arm_groups table is joined by equating the ‘arm_group_id’ to the ‘id’ column within the table. We are joining these extra tables here to further refine the set of clinical trials to only those where lenalidomide is used as an experimental therapy (as opposed to, for example, a comparator). Finally, when we join the clinial_trial_conditions table, we use two criteria: the ‘trial_id’ must match the ‘identifier’ of the trial in question and the ‘condition_id’ in the table must match that of the original orphan designation. This ensures that the returned trials match both the drug and condition of each designation.
One of the reasons why rare disease research is so difficult is rooted in the definition of rare diseases themselves: there are comparatively few patients with the condition. This can make running clinical trials exceptionally difficult and, in turn, increases the value of existing trial data to understand how previous trials were designed and their outcomes. Within clinicaltrials.gov, the reason for trial termination is simply a free text field. This is reasonable from the standpoint of a human reviewing individual trial data but makes it very difficult to understand the data at scale in a computational manner. With this in mind, we have introduced 30 standardized categories for why rare disease trials stop, including a flag capturing whether those reasons have to do with safety or efficacy. As an example, let’s find out why the trials for paclitaxel (DB01229), a microtubule inhibitor, have stopped:
SELECT ctwsc.category, ctwsc.definition, ctwsc.safety_efficacy_concern, COUNT(DISTINCT ct.identifier) AS 'Number of Trials' FROM orphan_designations od JOIN orphan_designation_drugs odd ON od.id=odd.orphan_designation_id JOIN clinical_trial_interventions_drugs ctid ON odd.drug_id=ctid.drug_id JOIN clinical_trial_interventions cti ON ctid.intervention_id=cti.id JOIN clinical_trial_intervention_arm_groups ctiag ON cti.id=ctiag.intervention_id JOIN clinical_trial_arm_groups ctag ON ctiag.arm_group_id=ctag.id JOIN clinical_trials ct ON cti.trial_id=ct.identifier JOIN clinical_trial_why_stopped_categories ctwsc ON ct.why_stopped_category_id = ctwsc.id WHERE odd.drug_id = 1229 AND ctag.kind = 'experimental' GROUP BY ctwsc.id ORDER BY COUNT(DISTINCT ct.identifier) DESC;Copy to clipboard

There are 18 rows in the output, the first six of which are shown here. Note the column ‘safety_efficacy_concern’ in the output. This is a boolean for whether the reason correlates with safety or efficacy concerns; here, it is set for “Safety” and “Efficacy,” but there are three other categories (“FDA Drug recall,” “MTD was Reached,” and “Safety - DSMB Restriction”) for which it applies. Here, the most common reason for trial termination was issues with patient recruitment; this turns out to be a common theme across all rare disease trials.
By taking unstructured text and placing it into standardized categories, we have made it much easier to compare trial termination reasons across trials and disease spaces. As an example, it is possible to narrow down all trial information to trials for a designated condition and understand the trial designs under which the trials experienced safety or efficacy concerns. This may be useful when designing future trials in these areas.
The query here is similar to the one used above for combining orphan designations with clinical trials but with two minor changes. Although it would be possible, and appropriate, to still match the orphan designation conditions to the clinical trials, it isn’t strictly necessary here since we are only matching trials that have had their termination reasons categorized. Cutting out some of these joins makes the query smaller and faster to run. The larger change is joining the results to the clinical_trial_why_stopped_categories table through the 'why_stopped_category_id' column on the main clinical_trials table (note that users who do not subscribe to the rare disease module will not have this column).
Rare diseases are currently underserved, often debilitating, conditions with few available treatment options. Orphan drug programs have provided incentives to accelerate research in these areas, but researchers still struggle with a lack of relevant information. Our rare disease dataset helps to bridge this gap when investigating drugs and conditions associated with FDA orphan drug designations. Both the drugs and conditions serve as entry points to other parts of our knowledgebase, such as structured indications. Furthermore, users can easily find matching trials and understand the existing clinical landscape for these underserved disease areas. In this manner, we provide rare disease researchers with a powerful set of tools to quickly gain knowledge and make informed decisions to spur their own research forward.